Chemical proteomics focuses on creating chemical tools and chemical probes tailored for the use in proteomics assays.
Protein-ABPP: Protein-based activity profiling using reactive covalent protein probes
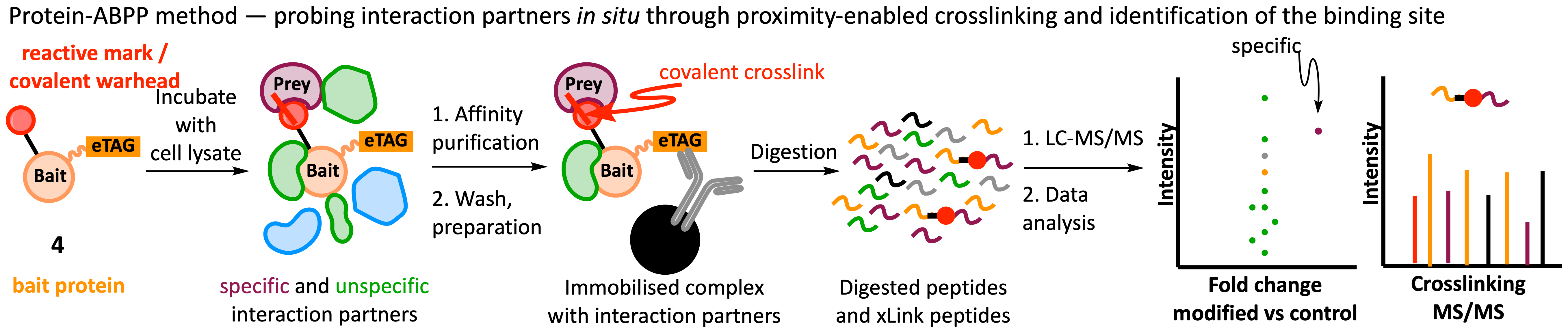
The recent discovery of methods for the incorporation of reactive amino acids into proteins through posttranslational chemical mutagenesis[1] has opened new possibilities for the study of protein-protein interactions and the investigation of molecular interactions. For this, reactive mimics of amino acids are incorporated in vitro using synthetic protein chemistry methods into target proteins. These mimics substitute either a canonical amino acid or an amino acid with a posttranslational modification such as acetylation, methylation, phosphorylation or glycosylation. Once incorporated, protein probes with reactive amino acid mimics can be used to covalently trap protein-protein interaction partners via proximity-enabled protein-protein crosslinking (PEPC). The reaction between the probe and its interaction partner is specific for the site of interaction (the binding site). Combined with a crosslinking mass spectrometry readout, these reactive protein tools allow to probe molecular interactions with amino-acid-level precision.
We develop tailored chemical proteomics methods in crosslinking mass spectrometry to be able to identify site-specific crosslinking events between reactive proteins and their interaction partners. We work on all aspects from sample preparation, data acquisition by high resolution mass spectrometry and data analysis.
Affinity matrices for the enrichment of subproteomes
We are starting projects developing affinity matrices to enrich for specific posttranslational modifications and proteomes, including:
- Protein O-glycosylation.
- Interaction partners for immunogenic antigens.
References
- Josephson B, Fehl C, Isenegger P, Nadal S, et al. “Light-driven post-translational installation of reactive protein side chains.” Nature 2020, 585, 530.
